This is a post about my OCR Performance Metric work for the Scribble Stadium product.
Currently, the Scribble Stadium product uses the Cloud Vision API to perform transcriptions but the plan is to move to a Tesseract-based transcription model.
In order to know when to move over to Tesseract we need to be able to compare the OCR performance of the Cloud Vision and Tesseract transcription engines. This post describes the effort to define the performance measure and build tooling to compute the measure.
The code for this project is available in https://github.com/Lambda-School-Labs/scribble-stadium-ds/tree/main/ocr_performance and the README file in the directory provides detailed info on the tools and their use.
The notebook with the data science work is at https://github.com/Lambda-School-Labs/scribble-stadium-ds/blob/main/TranscriptProcessing.ipynb
A blog post on this work has been published as an article on Medium - Scribble Stadium — Bridging the Analog and Digital.
The Reference dataset
The reference dataset consists of 4 sets of story files totalling 167 - 3101-3132, 3201-3248, 5101-5132, 5201-5264.
The associated transcript files are named <story number>.txt e.g. 3101.txt
If the story is contained in a single page, the associated image file will be <story number>.jpg e.g. 3101.jpg
If the story is spread over multiple pages, the associated image files will be <story number>-<page number>.jpg e.g. 3102-1.jpg,3102-2.jpg
There are a total of 167 transcript and 314 image files.
OCR performance metric
According to the Metrics for Complete Evaluation of OCR Performance paper, the basis for computing perf metrics is , essentially, aligning the hypothesis(ocr generated) transcripts and reference(ground truth) transcripts and using the Levenshtein distance between them at the character(asrtoolkit.cer) and word(asrtoolkit.wer) level, where the code for both these functions can be found in https://github.com/finos/greenkey-asrtoolkit/blob/master/asrtoolkit/wer.py.
The paper, in addition, points out that in the case of text-text comparisons where both the hypothesis and reference transcripts have no segmentation information, this approach works well for 1D(i.e. lines) texts but not 2D(i.e. pages) texts because of problems with aligning the texts.
In order to avoid the alignment problem, the proposed solution relies on applying NLP vectorization techniques and computing the CosineSimilarity between the vectors for the hypothesis and reference transcripts - we could also compute the distance between the vectors but it is not a very good measure with high dimensionality vectors and, besides, CosineSimilarity gives us a bounded value([0,1]) which makes comparisons easier.
Creating the Vectorizer and Dimensionality Reducer
- The first step is to load the
referencetranscripts
trex = rcompile('[^a-zA-Z 0-9]')
tokenize = lambda x: trex.sub('', x.lower().replace(',', ' ').replace('-', ' '))
def read_transcript(file_name):
file_names = glob(f'{file_name}*')
ret = ''
for fname in file_names:
with open(fname) as fd:
ret += fd.read()+'\n\n'
return ret
#Some of the reference transcripts have missing information and those files have
#filename with '(missing page <number>)' substrings - filter these files out for
#now.
rex = rcompile('[0-9]+.txt')
reference_transcripts_file_roots = [x.split('.')[0] for x in reference_transcripts_files if rex.match(x) is not None]
with Path(reference_transcripts_dir):
reference_transcripts = [read_transcript(f'{x}.txt') for x in reference_transcripts_file_roots]
reference_transcripts = list(map(tokenize, reference_transcripts))
- We then create and fit a
TfidfVectorizertoreference_transcripts
Instantiate vectorizer object
tfidf = TfidfVectorizer(stop_words='english',
ngram_range=(1,2),
min_df=1
)
# Create a vocabulary and get word counts per document
ref_dtm = tfidf.fit_transform(reference_transcripts)
features = tfidf.get_feature_names()
#display(len(features), features[:50])
# Get feature names to use as dataframe column headers
ref_dtm = pd.DataFrame(ref_dtm.todense(), columns=features)
- This gives us a
DTM(Document Term Matrix)where each document entry is a vector of length22296. We, therefore, train anAutoEncoderto build aDimensionality Reducer.
ishape = ref_dtm.shape[1] # 22296
# Create Model
input_img = Input(shape=(ishape, ))
x = Dense(512)(input_img)
x = Dense(256)(x)
x = Dense(128)(x)
encoded = Dense(64)(x)
x = Dense(128)(encoded)
x = Dense(256)(x)
x = Dense(512, activation='sigmoid')(x)
decoded = Dense(ishape, activation='sigmoid')(x)
rmodel = Model(input_img, decoded)
rmodel.compile(loss='mse', optimizer=Adam(learning_rate=0.01))
rmodel.fit(ref_dtm, ref_dtm, batch_size=512, epochs=5)
encoder = Model(input_img, encoded)
- We then use the dimensionality reducer
encoderto reduce the dimensionality of the previously computedDTM.
encoded_ref_dtm = encoder.predict(ref_dtm)
- The final step is to
pickleall the data that will be required to build the tooling
from joblib import dump
MODELS_DIR = '../models/'
TFIDF = MODELS_DIR+'tfidf.pkl'
ENCODED_REF_DTM = MODELS_DIR+'encoded_ref_dtm.pkl'
ENCODER = MODELS_DIR+'encoder.h5'
FILE_ROOTS = MODELS_DIR+'file_roots.pkl'
dump(tfidf, TFIDF)
encoder.save(ENCODER)
dump(encoded_ref_dtm, ENCODED_REF_DTM)
dump(reference_transcripts_file_roots, FILE_ROOTS)
CompareTranscriptions
The __init__ method loads in all the pickled objects from the previous section
# Load the model saved in encoder.h5
self.encoder = load_model(ENCODER)
# Load the TfIDF vectorizer saved in tfidf.pkl
self.tfidf = load(TFIDF)
# Load the encoded reference DTM saved in encoded_ref_dtm.pkl
self.encoded_ref_dtm = load(ENCODED_REF_DTM)
# Load transcript file roots list from file_roots.pkl
self.file_roots = load(FILE_ROOTS)
.compute_error(engine, hypothesis_transcripts_dir)
Given the location of the hypothesis transcripts(hypothesis_transcripts_dir), it
- loads the hypothesis transcripts
# Match characters that are not blank space, digits or
# letters in the alphabet.
rex = rcompile('[^a-zA-Z 0-9]')
def tokenize(x):
return rex.sub('', x.lower().replace(',', ' ').replace('-', ' '))
def read_transcript(file_name):
'''
Read and return the content of all transcript files
that have file_name as prefix.
e.g. A file name of .../google_transcripts/3102
would mean that the conents of
.../google_transcripts/3102-1
and
.../google_transcripts/3102-2
would be read and returned.
'''
# generate the list of all file names that match
# the wild carded file name
file_names = glob(f'{file_name}*')
ret = ''
for fname in file_names:
with open(fname) as fd:
ret += fd.read()+'\n\n'
return ret
with Path(transcripts_dir):
self.transcripts = [read_transcript(x) for x in self.file_roots]
self.transcripts = list(map(tokenize, self.transcripts))
- computes the dimensionality reduced
DTMfor thehypothesistranscripts
# Use the TfIDF vectorizer to vectorize the transcripts
dtm = self.tfidf.transform(self.transcripts).todense()
# Run the vectors through the encoder to reduce
# dimensionality
self.encoded_dtm = self.encoder.predict(dtm)
and
- finally computes the error measure for the
hypothesistranscripts based on the cosine similarity between the vectors for thereferenceand correspondinghypothesistranscripts.
# Convert cosine_similarity value to cos error
# cosine_similarity of 1 implies identical text
# so 1 minus that value is a measure of error.
def COS_ERROR(x): return abs(round((1-x)*1e7, 0))
cossim = []
for idx, ref in enumerate(self.encoded_ref_dtm):
cossim.append(
cosine_similarity([ref],
[self.encoded_dtm[idx]])[0][0])
# Convert cosine_similarity values into COS_ERROR values
# and store in coserr
coserr = [COS_ERROR(x) for x in cossim]
Computing performance metrics
Validate with Reference Transcriptions
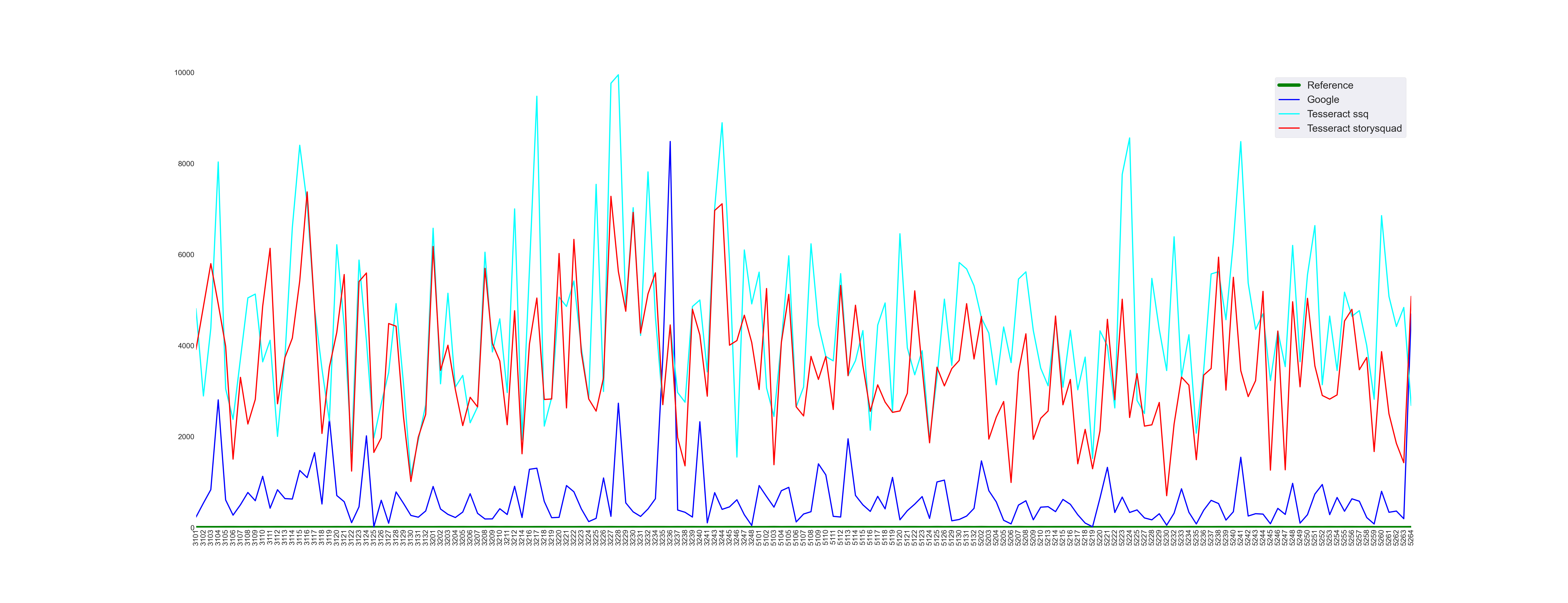
The first thing we need to do is to validate all the tooling by ensuring that we get an error value of 0 when we compare Reference transcripts to themselves. That works!
The one to beat - Google Transcriptions
The next step is to see how well Google’s Cloud Vision API performs. Running the tooling gives us a mean error value of 663.
Tesseract with LSTM model — small training set
The initial evaluation of the Tesseract engine used an LSTM model(named ssq) trained on a small subset of the available data. As expected its performance is much worse than Google’s, with a mean error value of 4428.
Tesseract with LSTM model — larger training set
We then evaluated Tesseract with an LSTM model(named storysquad) trained with a larger subset of the available data.
We expected a performance better than the one with the smaller training set and that’s what we did get, with a mean error value of 3567.
Visualizing performance metrics
To visualize the performance metrics, we
- load the
.csvfiles associated with the various runs
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
#DATA_DIR is set to location of all the error files
GOOGLE_COS_ERROR = DATA_DIR + 'google_cos_error.csv'
REFERENCE_COS_ERROR = DATA_DIR + 'ref_cos_error.csv'
TESSERACT_SSQ_COS_ERROR = DATA_DIR + 'tess_ssq_cos_error.csv'
TESSERACT_STORYSQUAD_COS_ERROR = DATA_DIR + 'tess_storysquad_cos_error.csv'
reference_error = pd.read_csv(REFERENCE_COS_ERROR)
google_error = pd.read_csv(GOOGLE_COS_ERROR)
ssq_error = pd.read_csv(TESSERACT_SSQ_COS_ERROR)
storysquad_error = pd.read_csv(TESSERACT_STORYSQUAD_COS_ERROR)
- draw line plots of each of the error sets
xticks = reference_error.index.to_list()
xlabels = reference_error.img_name.to_list()
sns.set(font_scale=4)
fig, ax = plt.subplots(1,1,figsize=(130,50), subplot_kw={'ylim': (0,10000), 'xlim':(0,164)})
ax.set_facecolor('white')
reference_error['ref_cos_error'].plot(ax = ax, linewidth=20,color='green', label='Reference');
google_error['google_cos_error'].plot(ax = ax, linewidth=7,color='blue', label='Google');
ssq_error['tess_ssq_cos_error'].plot(ax = ax, linewidth=7,color='cyan', label='Tesseract ssq');
storysquad_error['tess_storysquad_cos_error'].plot(ax = ax, linewidth=7,color='red', label='Tesseract storysquad');
plt.xticks(xticks, xlabels, rotation='vertical')
plt.legend(prop={'size': 60})
plt.savefig('cos_error_plot.png');
Here’s the resulting image